Author Summary CRISPR-Cas9 is a revolutionary biological technique for precisely editing cells’ genomes. Amongst its many capabilities is the deletion of defined regions of DNA, creating a wide range of applications from modelling rare human diseases, to performing very large knock-out screens of candidate regulatory DNA. CRISPR-Cas9 requires researchers to design small RNA molecules called sgRNAs to target their region of interest. A large number of bioinformatic tools exist for this task. However, CRISPR deletion requires the design of optimised pairs of such RNA molecules. This manuscript describes the first pipeline designed to accomplish this, called CRISPETa, with a range of useful features. We use CRISPETa to design comprehensive libraries of paired sgRNA for many thousands of target regions that may be used by the scientific community. Using CRISPETa designs in human cells, we show that predicted pairs of sgRNAs produce the expected deletions at high efficiency. Finally, we show that these deletions of genomic DNA give rise to correspondingly truncated RNA molecules, supporting the power of this technology to create cells with precisely deleted DNA.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|
|









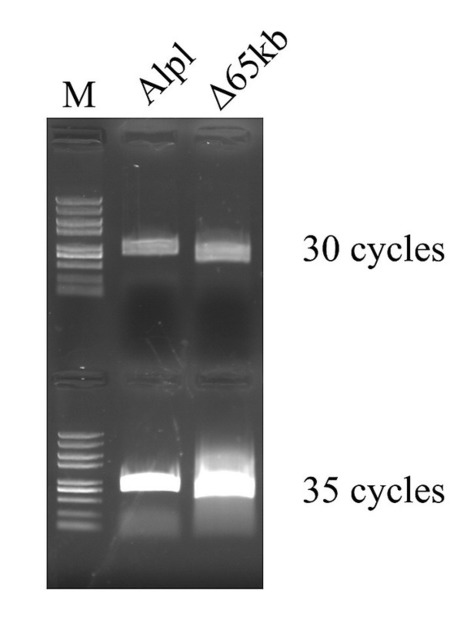





We the scientists present CRISPETa, a pipeline for flexible and scalable paired sgRNA design based on an empirical scoring model. Multiple sgRNA pairs are returned for each target, and any number of targets can be analyzed in parallel, making CRISPETa equally useful for focussed or high-throughput studies.